About the Lab
Main Direction: studying the molecular-genetic mechanisms that regulate morphogenesis.
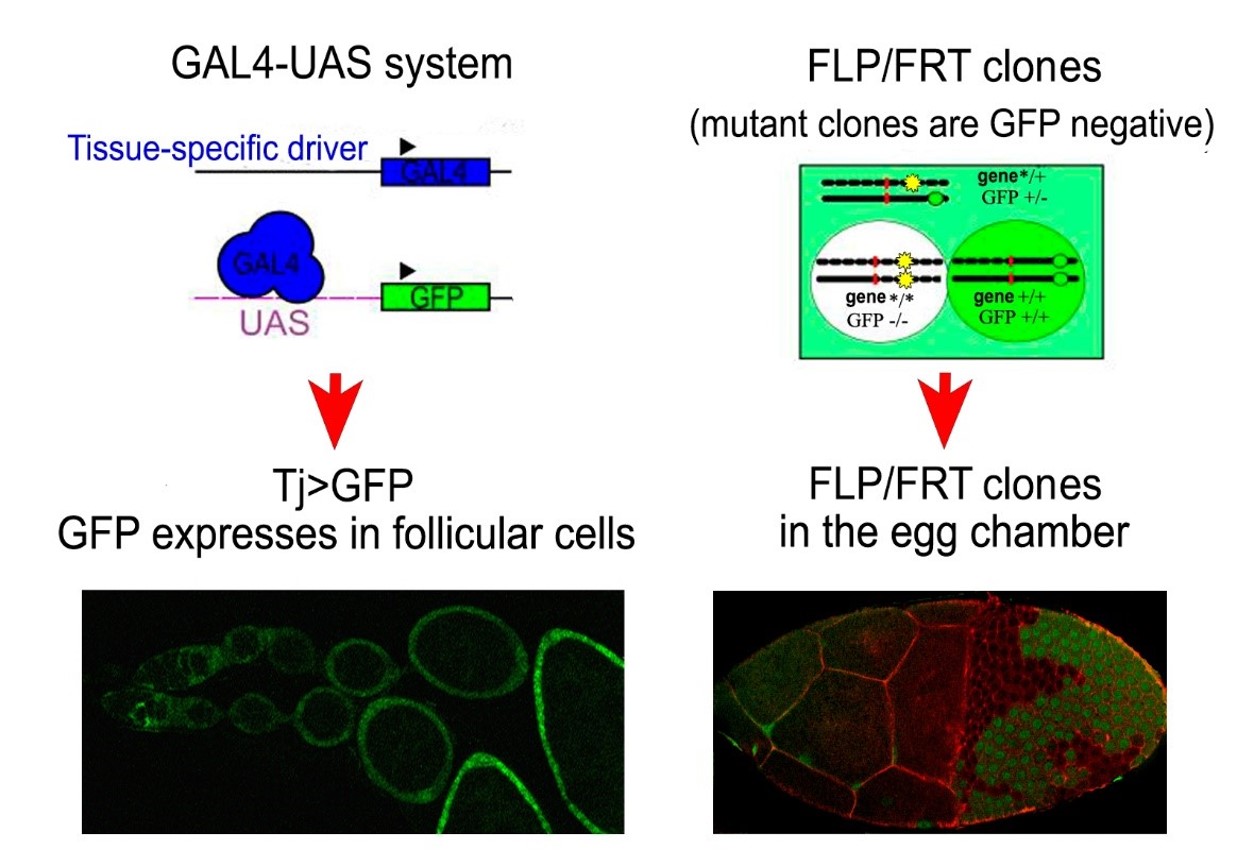
Fig.1 Tools for controlling the functions of genes in various tissues at various stages of development. The GAL4/UAS system contains yeast transcription activator GAL4 and the target gene controlled by the upstream activating sequence (UAS) (on the left). This system allowes us to modulate (down-regulate and over-produce) gene expression in living flies. The FLP/FRT system is used for the induction of mitotic clones (on the right). The FLP/FRT system uses FLP recombinase and target FRT-sites (highlighted in red) for the induction of homozygous mutant cell clones and wild type cells in heterozygous individuals. In the FLP/FRT system, heterozygous cells are labeled by one GFP copy (green background). After recombination, one of the daughter cells gets two copies of the mutant allele (yellow star) and no copies of the GFP-marker (green oval). Therefore, it has no green luminescence (white background). The other daughter cell gets two normal alleles of the studied gene and two copies of the GFP marker (dark green background). This system helps us to study homozygous mutant clones of cells, carrying alleles that cause adult lethality. Below are examples of the use of GAL4/UAS and FLP/FRT systems: specific expression of the reporter gene UAS-GFP under the control of Tj-GAL4 driver construct in the somatic cells of the female reproductive system in Drosophila (on the left); the mosaic egg chamber with mitotic clones induced by the FLP/FRT system (on the right). The clones are identified by the absence of green fluorescent protein (GFP–) luminescence.
The goal is to experimentally study the function of genes (protein domains, chromatin regions) and the genetic and epigenetic mechanisms responsible for their varying activity during development. We use model systems for the regulation of tissue-specific gene expression in vivo by targeted silencing or overexpression of genes (Fig. 1). To achieve this, we work with Drosophila melanogaster that has been transformed with fly, human and mouse genes. Some research is carried out in human and insect cell cultures.
We use various research methods, including bioinformatics analysis, classical genetic analysis, modern genetic engineering and embryo microinjection, immunohistochemistry, including immunostaining and confocal microscopy imaging, and biochemical analysis.
The research theme of the Laboratory includes tree sections.
Section 1: Antisense transcription and non-coding RNAs as potential sources of de novo gene birth and intergenic overlap creation.
The goal of this study is to understand the mechanism responsible for the coordinated expression of overlapping genes during ontogeny. How do overlapping genes arise in the eukaryotic genome? How is the overlapping region maintained despite the pressure of natural selection? How does co-expression occur in overlapping genes, and how does one gene influence the expression of another? What is the role of antisense transcription in this process? Recent bioinformatics studies have observed a trend towards emergence of overlaps between conserved and de novo gene. The search for and study of de novo genes presents a unique problem, which raises the question of how selective evolution can result in the emergence of functional genes in non-coding DNA. Research has demonstrated that a small reading frame with a promoter can be generated with only a few mutations. Nevertheless, the functionality of new short peptides that encode de novo genes is still unresolved. It is therefore relevant to ask how new genes and their products are integrated into existing gene networks and signaling pathways. Which mechanisms contribute to their adaptation?

Fig.3 The homeotic transformation of the antenna into the tarsus induced by the insertion of transposable P-element in the region of lawc-Trf2 (Simonova et al., 1992). The left image shows the wild phenotype, while the right image displays the mutant phenotype.
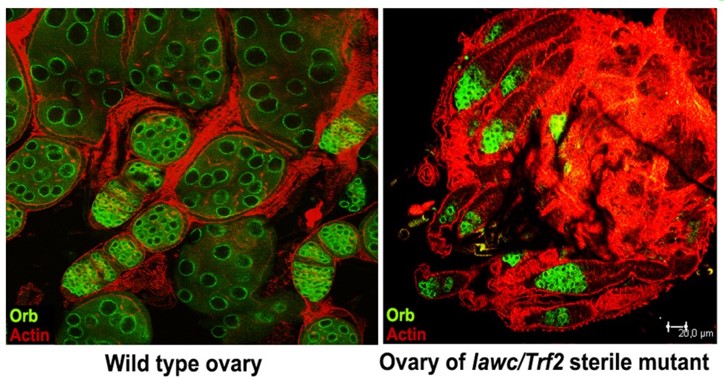
Fig.4 Immunofluorescent staining of the wild-type Drosophila ovary and the ovary of mutant lawc/Trf2 female. Actin cytoskeleton is stained with AlexaFluor phalloidin (red). The ovarian germ cells are labeled with anti-Orb antibodies (green). Note that the egg chamber formation has failed in the mutant ovary.

Fig.5 RNAi-mediated depletion of Trf2 leads to micronuclei formation and disturbs nuclear envelope in Drosophila S2 сells (Cherezov et al., 2020).
In our experiments, we use a model system of two overlapping Drosophila genes, one of which is the leg-arista-wing complex (lawc) gene, which arose de novo, and the other encodes a conserved general transcription factor, TBP-related factor 2 (Trf2) (Fig. 2).
In previous experiments, we discovered that mutations in lawc-Trf2 can result in the homeotic transformation of the antenna into the tarsus (Fig. 3), cause the female sterility (Fig. 4), and change the chromosome structure. The depletion of Trf2 disrupts the integrity of the nucleus in cells and leads to micronuclei formation (Fig. 5). The function of the lawc gene is not well understood.
Section 2: Significance of neuron-specific, evolutionarily conserved D4 family transcription factors in pathways that regulate morphogenesis.
A group of novel neuro-specific genes of the evolutionarily conserved d4 family was first identified and characterized in collaboration with Prof. Vladimir Buchman (Cardiff University, UK). The consequences of dysfunction of two mouse neurogenes, neuro-d4 and cer-d4, were first investigated. The d4 gene products (PHD-type zinc fingers, DPF) are thought to be involved in the epigenetic regulation of neurogenesis. The existing of several d4 paralogs with overlapping expression, the complexity of the splicing pattern cause difficulties in the study of this family genes in mammals. We are currently studying the function of d4 genes, toothrin and drosophila d4, in Drosophila. The study is being carried out by means of genetic, immunohistochemical and biochemical methods. We plan to obtain and characterize null mutants of toothrin and drosophila d4 genes; obtain Drosophila lines with the expression of TTH and DD4 proteins cross-linked with fluorescent markers, and create a system for targeted suppression of the expression of these genes in vivo (Fig. 6).

Fig.6 Localization of tth gene within D. melanogaster genome, highlighted in red (a), and the designing of the construct for the expression of TTH:GFP protein (b) (Kuvaeva et al., 2022).
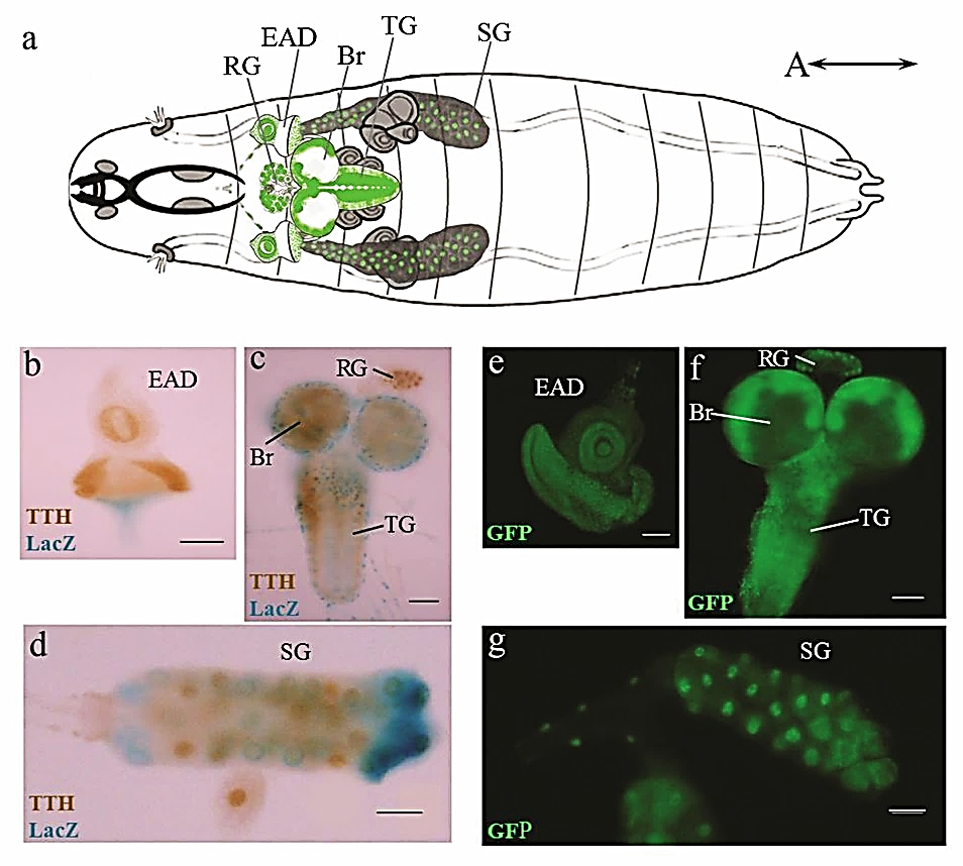
Fig.7 Pattern of TTH expression at the larval stage of Drosophila development. (a) schematic representation; (b - d) immunohistochemical staining with antibodies against TTH (brown), LacZ expression in glial cells under the Repo promoter (blue); (e - g) the localisation of chimeric protein TTH::GFP in the tissues of the larva. RG - ring gland, EAD - eye-antennal imaginal disc, Br - brain, TG - thoracic ganglion, SG - salivary gland. The scale bar - 50µm (Kuvaeva et al., 2022).
Preliminary data indicate TTH involvement in the visual system's development and function, as well as the neurohumoral regulation of morphogenesis (Fig. 7). We have planned a series of behavioral tests to clarify how the epigenetic neuronal D4 family factors impact the function of the sensory systems.
Section 3: Genetic mechanisms regulating both morphogenesis and biodegradation of xenobiotics.
Aryl hydrocarbon receptor, AHR, is a ligand-dependent transcription factor associated with xenobiotic detoxification and carcinogenesis. AHR ligands include substances that are part of pharmaceuticals, including those used in oncotherapy. Current work is to study the Aryl hydrocarbon receptor signaling pathway in the biodegradation of pharmacological agents in norm and pathology, in the maintenance of homeostasis and in the processes of cell differentiation. We use both transgenic Drosophila lines carrying human AHR for the study of AHR functions in vivo and human cancer cell lines for the study of its role in carcinogenesis.

Fig.8 Effects of ligand-depended human AHR activation induced in Drosophila eye imaginal discs. Eye phenotypes of GMR>AHR flies developed on standard medium without exogenous ligands (A, A′), on medium with indinol (B, B′), beta-Naphthoflavone (C, C′) or indirubin (D, D′). Ommatidia are arranged in a highly regular pattern in control flies (A–A′), while flies reared on medium with exogenous ligands develop roughened eye phenotypes with irregular pattern and decreased number of mechanoreceptors (B–D, B′–D′) (Akishina et al., 2017).

Fig.9 A humanized test line of Drosophila transformed with the human AHR gene designed to assess the effects of xenobiotics on the development of evolutionarily conserved organ and tissue structures.
Using Drosophila as a model to assess function of the human AHR. In contrast to human AHR, Drosophila AHR could not be activated by exogenous ligands. This allows us to study the consequences of tissue-specific ligand-depended human AHR activation in transgenic flies (Fig. 8).
Moreover, we have developed and patented a method for assessing the pharmacological and toxic properties of substances - potential human AHR ligands (Fig. 9).
The highly conservative human AHR signaling pathway provides an opportunity to examine its in vivo functions through the use of transgenic Drosophila melanogaster with inducible human AHR expression. In previous studies, we found that transcriptional regulation by AHR is linked to developmental activities of epigenetic modulators of chromatin structure. In other words, we demonstrated that not all AHR target genes may be activated in response to AHR ligand treatment. Its activity depends on the chromatin architecture of the target gene regulatory regions. The tight chromatin may impede the accessibility of the DNA binding sites to the AHR/ARNT transcription complex (Fig. 10).

Fig.10 Genetic depletion of Рс (A) and inhibition of E(z) H3K27me3 activity by UNC1999 (B) leads to an increase in the expression of some AHR target genes using ectopic expression of the human AHR and some of its exogeneous ligands (Akishina et al., 2017).
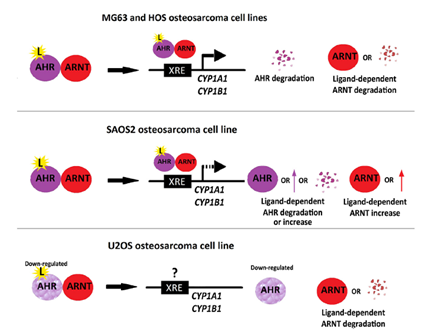
Fig.11 Models describing the AHR/CYP1 signaling pathway and its dysregulations found in MG63, HOS, SAOS2, and U2OS osteosarcoma cells. In MG63 and HOS cells, CYP1A1 and CYP1B1 gene expression is induced, and AHR protein and ARNT protein levels are decreased in response to the ligands (L). (Vorontsova et al., 2023).
Since exogenous AHR ligands and small molecule inhibitors of epigenetic modifiers are often used as pharmaceutical anticancer drugs, our findings may have significant implications in designing new combinations of therapeutic treatments for oncological diseases.
The Aryl hydrocarbon receptor/Cytochrome 450 signaling pathway in osteosarcoma cell lines. Osteosarcoma is the most common malignant tumor of bone, with rapid progressive growth, early distant metastases, and frequent recurrence after surgical treatment. Osteosarcoma is characterized by changes in the ratio and expression of different cytochrome P450 (CYP) isoforms that can affect the effectiveness of anticancer therapies. The expression of CYP1 genes is controlled by the ligand-dependent activity of AHR. We examined the AHR/CYP1 signaling pathway in four osteosarcoma cell lines (MG63, HOS, SAOS2, and U2OS) induced by the known AHR ligands: indirubin, indole-3-carbinol, and beta-naphthoflavone. Using qPCR and Western blot analysis, we explored the effects of these ligands on the expression of the CYP1 genes and studied the correlation between these responses and the changes in the mRNA and protein levels of AHR and the AHR nuclear translocator (ARNT) in these osteosarcoma cell lines. The results show that the AHR/CYP1 signaling pathway retains its function only in MG63 and HOS cells, and is impaired in SA OS2 and U2OS cells. Our data should be taken into account when recommending new strategies for the treatment of osteosarcoma and when evaluating new drugs against osteosarcoma in vitro (Fig. 11).
Lab members
Olga Simonova
Head of the Lab, PhD, Doctor of Biological Sciences
Reseacrh interests: developmental genetics, transcriptional factors, overlapping genes and antisense transcription, epigenetics, gene evolution, Drosophila.
Selected publications
1. Kuvaeva E.E., Kulikova D.A., Simonova O.B. and Mertsalov I.B. Generation of a null mutation of the toothrin gene by targeted homologous recombination in Drosophila melanogaster // Rus J of Dev Biol. – 2023. - Vol. 54. - No. 3. - P. 210–215. DOI: 10.1134/S1062360423030050
2. Vorontsova J.E., Akishina A.A., Cherezov R.O., Simonova O.B. A new insight into the Aryl Hydrocarbon Receptor/Cytochrome 450 signaling pathway in MG63, HOS, SAOS2, and U2OS cell lines // Biochimie. – 2023. - V. 207. - P. 102-112 - DOI: https://doi.org/10.1016/j.biochi.2022.10.018
3. Kuvaeva E.E., Kulikova D.A., Simonova O.B. and Mertsalov I.B. Studying the specific localization of Toothrin protein from related D4 family in Drosophila melanogaster // Rus J of Dev Biol. – 2022. - Vol. 53. - No. 2. - P. 145–149. DOI: 10.1134/S1062360422020072
4. Akishina A.A., Cherezov R.O., Vorontsova Y.E., Simonova O.B. Effect of the histone deacetylase (HDAC) inhibitor belinostat on the expression of the aryl hydrocarbon receptor and its target genes in human cell cultures // Rus J Dev Biol. – 2022. – Vol. 53. – № 2. – P. 134–137. DOI: 10.1134/S1062360422020023
5. Cherezov R.O., Vorontsova Ju.E., and Simonova O.B. The phenomenon of evolutionary “de novo generation” of genes // Rus J Dev Biol. – 2021. - Vol. 52. - No. 6. - P. 395–405. DOI: 10.1134/S1062360421060035
6. Zatsepina O.G., Nikitina E.A., Shilova V.Y., Chuvakova L.N., Sorokina S., Vorontsova J.E., Tokmacheva E.V., Funikov S.Y., Rezvykh A.P. & Evgen’ev M.B. Hsp70 affects memory formation and behaviorally relevant gene expression in Drosophila melanogaster // Cell Stress and Chaperones. 2021. - V.26 (3). – P. 575-594 https://doi.org/10.1007/s12192-021-01203-7
7. Rezvykh A.P., Funikov S.Y., Protsenko L.A., Kulikova D.A., Zelentsova E.S., Chuvakova L.N., Blumenstiel J.P., Evgen’ev M.B. Evolutionary dynamics of the pericentromeric heterochromatin in Drosophila virilis and related species // Genes. -2021. - 12(2). - P. 175. https://doi.org/10.3390/genes12020175
8. Cherezov R.O., Vorontsova J.E., Simonova O.B. TBP-related Factor 2 as a trigger for Robertsonian translocations and speciation // Int. J. Mol. Sci. 2020. V.21 (22). 8871. doi:10.3390/ijms21228871
9. Vorontsova J.E., Akishina A.A., Cherezov R.O., Simonova O.B. Functional activity of aryl hydrocarbon receptor in human osteosarcoma cell cultures // Moscow University Biological Sciences Bulletin. – 2020. - vol. 75. – No 4. – pp. 247–251. DOI: 10.3103/S0096392520040136
10. Shirokova A.V., Volovik V.T., Zagoskina N.V., Zaitsev G.P., Khydyakova H.K., Korovina L.M., Krutius O.N., Nikolaeva T.N., Simonova O.B., Alekseev A.A. Baranova E.N. From Dimness to Gloss – Characteristics of the Spring Rapeseed Mutant Form without Glaucous Bloom (Brassica napus L.) // Agronomy-Basel. – 2020. – Vol. 10. – Art. No 1563. DOI:10.3390/agronomy10101563.
11. Akishina A.A., Kuvaeva E.E., Vorontsova Y.E., and Simonova O.B. NAP family histone chaperones: characterization and role in ontogenesis // Rus J of Dev Biol. – 2020. - Vol. 51. - No. 6. - pp. 343–355. DOI: 10.1134/S1062360420060028
12. Funikov S., Rezvykh A., Kulikova D., Zelentsova E., Protsenko L., Chuvakova L, Tyukmaeva V., Arkhipova I., Evgen’ev E. Adaptation of gene loci to heterochromatin in the course of Drosophila evolution is associated with insulator protein // Scientific Reports. – 2020. – Vol. 10(1). – Art. no. 11893. DOI: 10.1038/s41598-020-68879-2.
13. Yurinskaya M.M., Krasnov G.S., Kulikova D.A., Zatsepina O.G., Vinokurov M.G., Chuvakova L.N., Rezvykh A.P., Funikov S. Y., Morozov A. V. & M. B. Evgen’ev H2S counteracts proinflammatory effects of LPS through modulation of multiple pathways in human cells // Inflamm. Res. 2020. V. 69. P. 481–495 https://doi.org/10.1007/s00011-020-01329-x
14. Vorontsova J. E., Zavoloka E.L., Cherezov R.O., Simonova O.B. Three important discoveries in the field of the cytoskeleton’s proteins functioning on the Drosophila melanogaster model // Molecular Biology, 2019, Vol. 53, No. 1, pp. 1–12. DOI: 10.1134/S0026893319010163
15. Akishina A.A., Vorontsova J.E., Cherezov R.O., Slezinger M.S., Simonova O.B., Kuzin B.A. NAP family CG5017 chaperone pleiotropically regulates human AHR target genes expression in Drosophila testis // Int. J. Mol. Sci. 2018. V. 20(1): 118. doi: 10.3390/ijms20010118
16. Vorontsova J.E., Cherezov R.O., Kuzin B.A., Simonova O.B. Aryl-hydrocarbon receptor as a potential target for anticancer therapy // Biochemistry (Moscow) Supplement Series B: Biomedical Chemistry. 2019. V. 13. no. 1. P. 36-54. DOI: 10.1134/S1990750819010116
17. Akishina A.A., Vorontsova J.E., Cherezov R.O., Mertsalov I.B., Zatsepina O.G., Slezinger M.S., Panin V.M., Petruk S., Enikolopov G.N., Mazo A., Simonova O.B., Kuzin B.A. Xenobiotic-induced activation of human Aryl hydrocarbon receptor target genes in Drosophila is mediated by the epigenetic chromatin modifiers // Oncotarget. 2017. V.8. No.61. P.102934-102947. doi.org/10.18632/oncotarget.22173
18. Vorontsova J., Cherezov R. and Simonova O. The Effect of TBP related factor 2 on chromocenter formation and chromosome segregation in Drosophila melanogaster // Chromosomal Abnormalities - A Hallmark Manifestation of Genomic Instability, IntechOpen, 2017. Chapter 8. P. 145-159. Edited by M.L. Larramendy and S. Soloneski. 202 pages. ISBN 978-953-51-3474-9. Print ISBN 978-953-51-3473-2. DOI: 10.5772/67314.
19. Kuzin B.A., Nikitina E.A., Cherezov R.O., Vorontsova J.E., Slezinger M.S., Zatsepina O.G., Simonova O.B., Enikolopov G.N., Savvateeva-Popova E.V. Combination of hypomorphic mutations of the Drosophila homologues of Aryl hydrocarbon receptor and Nucleosome assembly protein family genes disrupts morphogenesis, memory and detoxification // PloS One. 2014. V.9. №4: e94975. doi:10.1371/journal.pone.0094975
20. Cherezov R.O., Simonova O.B. Overlapping Genes and Antisense Transcription in Eukaryotes // Rus. J. Genet. 2014. Vol. 50, No. 7, pp. 653–666. DOI: 10.1134/S1022795414070059
21. Kulikova D.A., Mertsalov I.B., Simonova O.B. d4 family genes: Genomic organization and expression // Rus. J. Dev. Biol. 2013, V. 44, Issue 1, pp 1-6. DOI: 10.1134/S1062360413010037
22. Cherezov R.O., Vorontsova Yu.E., Mertsalov I.B., Kulikova D.A., Simonova. The influence of hairpin RNA against lawc, on the expression of overlapping genes of the lawc/Trf2 complex in D. melanogaster // Biology Bulletin, 2013, Vol. 40, No. 2, pp. 119–123. DOI: 10.1134/S1062359013020040
23. Vorontsova Yu.E., Cherezov R.O., Simonova. Effect of mutations in lawc/Trf2 gene on development of chromocenter and chromosome disjunction in Drosophila melanogaster // Rus. J. Genet. 2013. Vol. 49, No. 6, pp. 577–587. DOI: 10.1134/S1022795413050128
24. Simonova O.B., Modestova E.A., Vorontsova J.E., and Cherezov R.O. Screening of genomic regions affecting lawc/Trf2 gene expression during Drosophila melanogaster development // Rus. J. Dev. Biol. 2012. Vol. 43. No. 5. pp. 301–317. 10.1134/S1062360412050086
25. Vorontsova Yu.E., Cherezov R.O., Zatsepina O.G., Slezinger M.S., Kuzin B.A., Simonova O.B. Gene expression modulation is an evolutionary resource of adaptive alterations in the morphogenesis of insect limbs // Biology Bulletin, 2012, Vol. 39, No. 2, pp. 186–193. DOI: 10.1134/S1062359012020124
26. Kuzin BA, Modestova EA, Vorontsova IuE, Zatsepina OG, Mikaelian AS, Slezinger MV, Simonova OB. Interaction of the ss and CG5017 genes in the regulation of morphogensis of limbs in Drosophila melanogaster // Rus. J. Dev. Biol. 2010 Sep-Oct;41(5):364-9. Russian DOI: 10.1134/S1062360410050061
27. Simonova O.B. and Burdina N.V. Morphogenetic movement of cells in embryogenesis of Drosophila melanogaster: mechanism and genetic control // Rus. J. Dev. Biol. 2009. 39, 283–299 DOI: 10.1134/S1062360409050038
28. Simonova OB, Vorontsova IuE. Source of asymmetry in ontogeny: early polarization of the germline cyst and oocyte in Drosophila // Genetika. 2008. 44(9):1157-71. Review. Russian. DOI: 10.1134/S1022795408090019
23. Vorontsova J.E., Modestova E.A., Burdina N.V., Korochkin L.I., Simonova O.B. Restoring viability of lethal mutants for the leg-arista-wing-complex gene in rescue experiments with transgenic constructs that express the trf2 gene domains in Drosophila melanogaster // Dokl. Biol. Sci. 2007. Т. 417. № 1. P. 429-431. DOI: 10.1134/S0012496607060051
24. Kopytova D.V., Krasnov A.N., Kopantceva M.R., Nabirochkina E.N., Nikolenko J.V., Maksimenko O., Kurshakova M.M., Lebedeva L.A., Yerokhin M.M., Simonova O.B., Korochkin L.I., Tora L., Georgiev P.G. and Georgieva S.G. Two isoforms of Drosophila TRF2 are involved in embryonic development, premeiotic chromatin condensation and proper differentiation of germ cells of both sexes // Mol. Cell Biol. 2006. V.26. № 20. P. 7492 – 505
25. Modestova E.A., Vorontsova Y.E., Korochkin L.I., and Simonova O.B. Induction of lethal mutations of the leg-arista-wing complex gene of Drosophila melanogaster // Doklady biological sciences. 2005. V.403. PP. 282-283.
26. Kopytova D.V., Krasnov A.N., Simonova О.B., Modestova Е.А., Korochkin L. Georgieva S.G. Study of the lawc-trf2 gene of Drosophila melanogaster and the protein product of this gene // Dokl Biochem Biophys. 2005. V.405. PP. 380-382.
27. Simonova О.B., D.A. Kulikova, I.B. Mertsalov, O.N. Umnova, V.N. Bashkirov, V.L. Buchman, L.I. Korochkin. Analysis of overexpression of thoothrin gene in Drosophila melanogaster // Genetics (Russia). 2005. V.41. №2. P. 138-143.
28. Modestova Е.А., Kopytova D.V., Georgieva S.G., Simonova О.B. P-Ph-mediated repression of leg-arista-wing complex gene transcription in Drosophila // Genetics (Russia). 2003. №5. P.713-716.
29. Nabirochkina E., Simonova O.B., Mertsalov I.B., Kulikova D.A., Ladigina N.G., Korochkin L.I. and Buchman V.L. Expression pattern of dd4, a sole member of the d4 family of transcription factors in Drosophila melanogaster // Mech Dev. 2002. V. 114. PP.119-123.
30. Ninkina N.N., Mertsalov I.B., Kulikova D.A., Alimova-Kost M.V., Simonova O.B., Korochkin L.I., Kiselev S.L., Buchman V.L. Cerd4, third member of the d4 gene family: expression and organization of genomic locus // Mammalian Genome. 2001. V. 12. PP.862-866.
31. Zakharenko L.P., Gracheva E.M., Romanova O.A., Zakharov I.K., Voloshina M.A., Kochieva E.Z., Simonova O.B., Golubovsky M.D., Georgiev P.G. hobo-induced rearrangements are responsible for mutation burst at yellow locus in a natural population of Drosophila melanogaster // Mol. Gen. Genet. 2000. V. 263. PP. 335-341.
32. Simonova O.B. A novel trans-regulatory locus of Drosophila // Genetics (Russia). 2000. V.36. N11. P. 1464 – 1474.
33. Simonova O.B., Sukhoverkhova T.I., Zhuravel D.A., Romanova L.G., Korochkin L.I. A study of olfactory response in Drosophila melanogastr leg-arista-wing complex homeotic mutants // Genetics (Russia). 2000. V.36. N11. P. 1531 – 1534.
34. Simonova O.B., Sukhoverkhova T.I., Jansugurova L.B., Korochkin L.I. Taste perception analysis in the leg-arista-wing complex homeotic mutants of Drosophila // Genetics (Russia). 2000. V.36. N5 P.657 – 665.
35. Mertsalov I.B., Kulikova D.A., Ninkina N.N., Simonova O.B., Buchman V.L., Korochkin L.I. Genomic organization of the mouse neuro-d4 gene // Genetics (Russia). 2000. V.36. N3 P.314 – 317.
36. Kulikova D.A., Mertsalov I.B., Ninkina N.N., Simonova O.B., Buchman V.L., Korochkin L.I. Genomic organization of the mouse ubi/requem-d4 gene // Doklady biological sciences. (Russia). 2000.
37. Petruk S. F., Jagaeva I.V., Soldatov A.V., Simonova O.B. Cloning of the leg-arista-wing complex (lawc) gene and characterization of its mutant derivatives in Drosophila // Genetics (Russia). 1998. V.34. N.3. P. 446-448.
38. Simonova O.B., Petruk S. F., Jagaeva I.V., Korochkin L.I. The role of lawcp1 mutation in the regulation of white gene expression in Drosophila // Genetics (Russia). 1998. V.34. N.3. P. 349-354.
39. Simonova O.B., Petruk S.F. and Korochkin L.I. Genetic analysis of combinations of the lawcp1 mutation with various alleles of AS-C genes in Drosophila // Genetics (Russia). 1996. V.32. N.7. P. 949-955.
40. Simonova O.B., Petruk S.F. and Korochkin L.I. Differential characteristics of the temperature-sensitive period of the lawcp1 mutation based on macrochaeta overexpression in Drosophila // Genetics (Russia). 1996. V.32. N.3. P. 445-447.
41. Simonova O.B., Petruk S.F., Gerasimova T.I. and Korochkin L.I. Determination of the temperature-sensitive period of a new mutation lawcp1 in Drosophila melanogaster // Genetics (Russia). 1995. V. 31. N. 9. P.1243-1248.
42. Gerasimova T.I., Gdula D., Gerasimov D. V., Simonova O.B., Corces V. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position -effect variegation // Cell. 1995. V. 82. P. 587-597.
43. Simonova О.B., Кuzin B.А., Georgiev P.G., Gerasimova Т.I. Novel regulatory mutation of Drosophila melanodaster // Genetics (Russia). 1992. N2. V.28. P.164-167.
44. Kuzin B.А., Dosjаnоv К.Т., Simonova О.B., Gerasimova Т.I., Gulyaev D.V. A novel allelic ssa variant and its action in regulation of leg and antennal discs proliferation in Drosophila melanodaster // Ontogenez (Russia). 1991. N2. V22. P.212-216.
45. Georgiev P.G., Kiselev S.L., Simonova O.B., Gerasimova T.I. A novel transposition system in Drosophila melanogaster depending on the Stalker mobile genetic element // EMBO J. 1990. V. 9. N. 7. P.2037-2034.
46. Georgiev G.P., Tchurikov N.A., Ilyin Yu. V., Georgieva S.G., Mizrokhi L.Y., Priimagii A.F., Gerasimova T.I., Georgiev P.G., Simonova O.B., Kiselev S.L., Kochieva E.Z. Mobile genetic elementsin Drosophila melanogaster // Genome. 1989. V. 31. N 2. P. 920-928.












