About the Lab
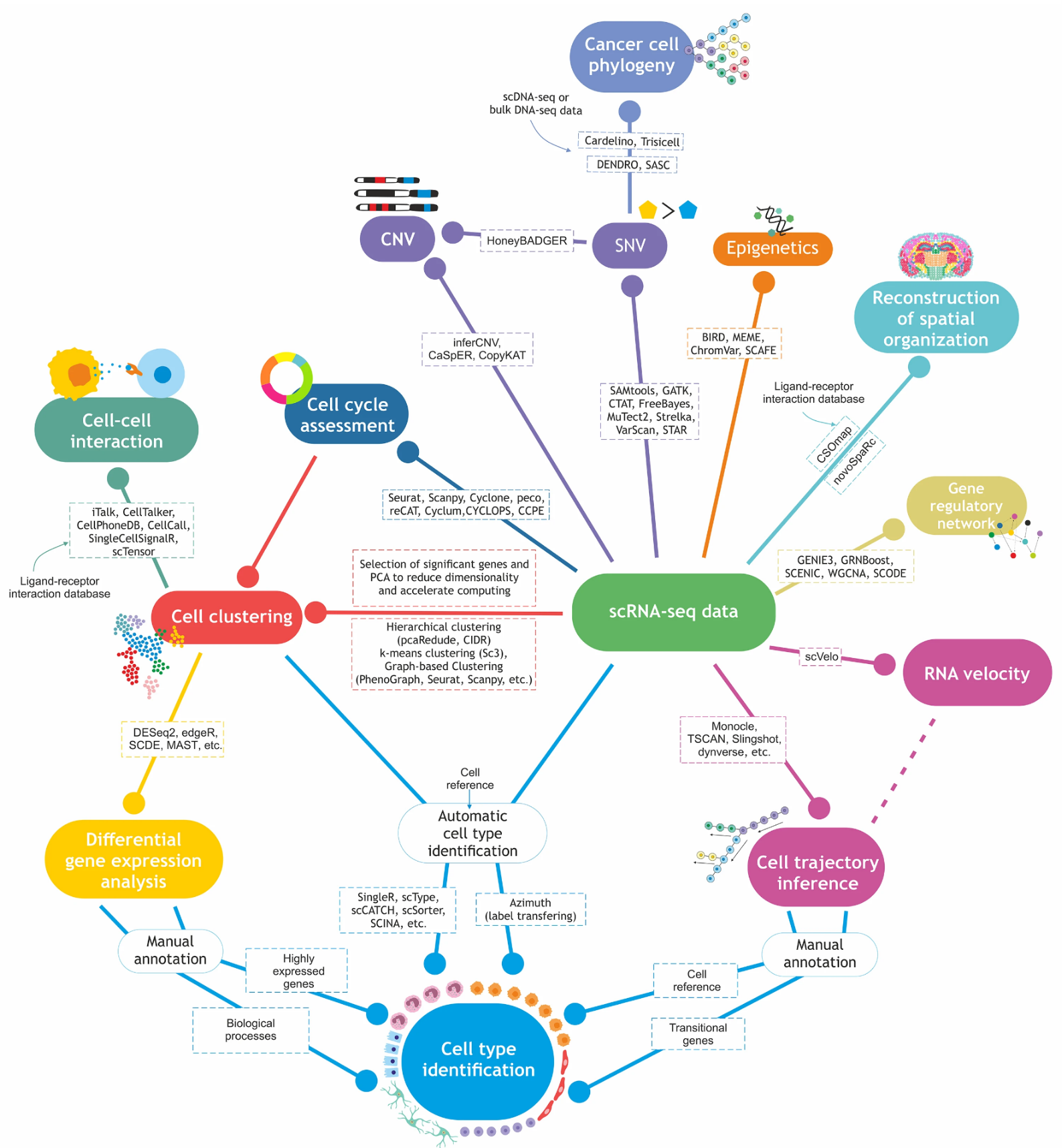
Fig. 1 Current approaches to analyzing single cell RNA sequencing data (Khozyainova et al, Biochemistry (Moscow), 2023)
The laboratory was established in 2022 within the framework of the project # 075-15-2021-1075 supported by the program "Genetic Technologies" of the Russian Ministry of Education and Science. The main direction of our research is the examination of molecular and cellular organization of the brain and sensory systems in the context of developmental disorders of the nervous system, metabolic, endocrine and psychiatric diseases and neurodegenerative disorders.
The major experimental approach employed in the lab, is single cell RNA sequencing (scRNA-seq) of the brain region of interest. A key advantage of this approach is the ability to study gene expression in each individual cell in the sample, allowing us to gain insight into gene activity at a fundamentally different resolution in comparison to previous studies. The most important step of the work is the analysis of the sequencing data. The analysis employs the entire broad arsenal of computational methods available, which have been developed specifically to analyze this type of data.
In addition to off-the-shelf software, our team is working on the development and optimization of algorithms and tools for single cell sequencing data analysis.
Models of type 1 and type 2 diabetes
Nowadays, type 1 and type 2 diabetes have emerged as socially significant diseases, posing serious challenges. Approximately 8.8% of the global adult population is diagnosed with diabetes, consequently giving rise to a plethora of severe metabolic disorders and obesity, specifically in the case of type 2 diabetes. The widespread prevalence of this issue places immense strain on the overall healthcare system due to the heightened risk of comorbidities. Previously established evidence has underscored the pivotal role of the hypothalamus in regulating metabolic processes associated with diabetes-related disorders, particularly in terms of weight control and maintaining stable endocrine gland function.
To induce diabetes, we administer injections of streptozotocin to neonatal animals, wherein a dosage of 150 mg/kg is used for the creation of the type 1 diabetes model, and 30 mg/kg for the establishment of the type 2 diabetes model. The focal point of our research resides in the hypothalamus, as it represent the neuroendocrine hub within the brain. Under experimental conditions, we analyze alterations in cell phenotypes within this specific region of the brain.
Data analysis based on the results from single cell RNA sequencing (scRNA-seq) conducted on both control and experimental animal groups. Additionally, we incorporate pertinent information derived from published papers within the realm of this research topic to complement our findings.
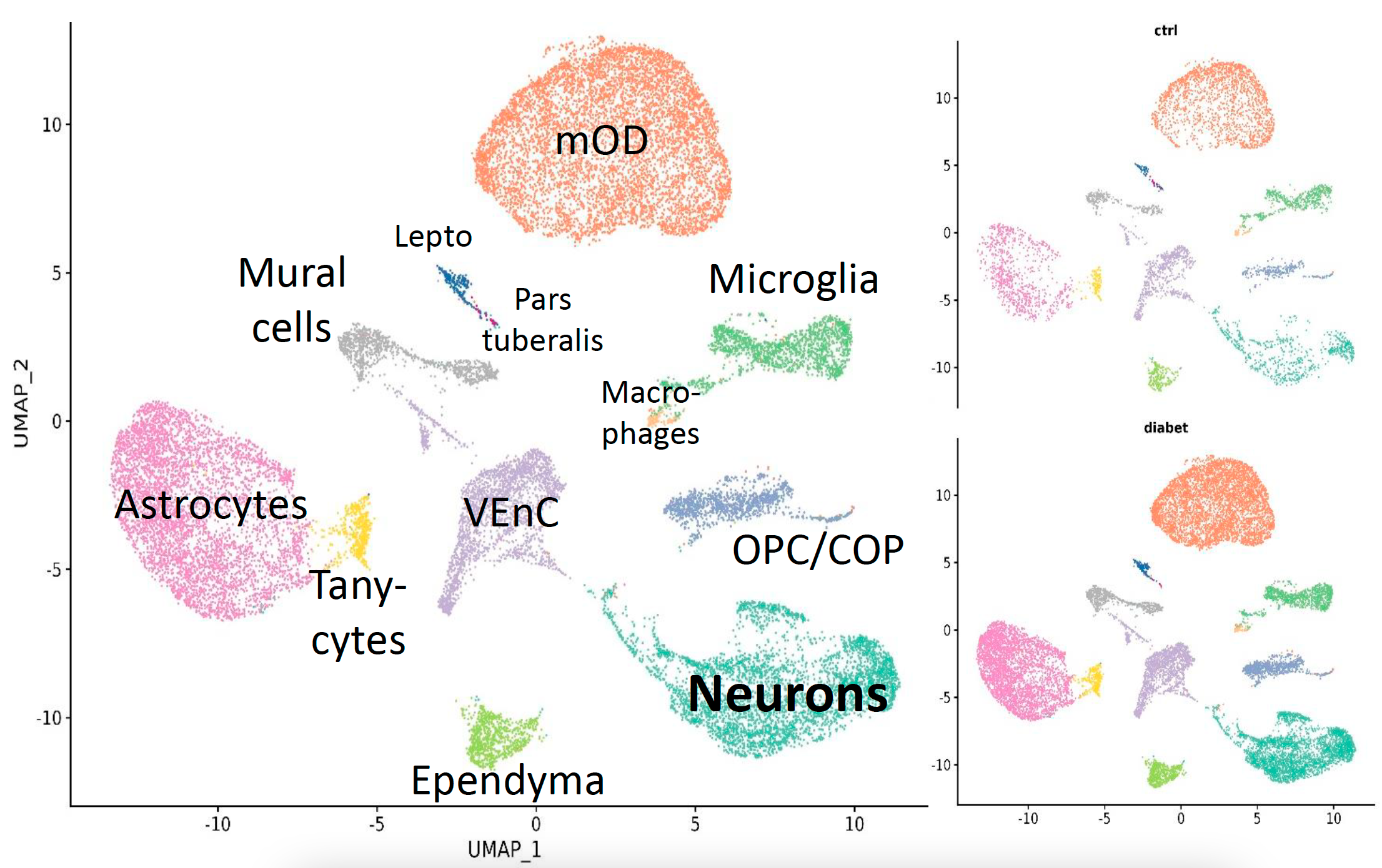
Fig. 2 Cell types composition in the hypothalamic cell sequencing dataset in type I diabetes among control and stimulated samples.
Transcriptomic changes during fear conditioning
The amygdala, an integral component of the limbic system within the brain, occupies a crucial position in the domain of emotion formation, particularly fear. Moreover, it actively engages in processes associated with memory consolidation and learning. These functions substantiate the amygdala's significance in the etiology of anxiety disorders and its substantial involvement in anxiety manifestations observed in various autism spectrum disorders, which ultimately impairs the overall quality of life for affected individuals, especially with regard to social integration, particularly in children. While the mechanisms of synaptic plasticity triggered by fear learning have been extensively elucidated and continue to be subject to investigation, a comprehensive characterization of the amygdala transcriptome at the single cell level in fear learning models has not yet been achieved. By scrutinizing transcriptional changes elicited by stress and fear induction in animals, we aim to identify the mechanisms of synaptic plasticity specific to distinct cell types, thereby filling this crucial knowledge gap.
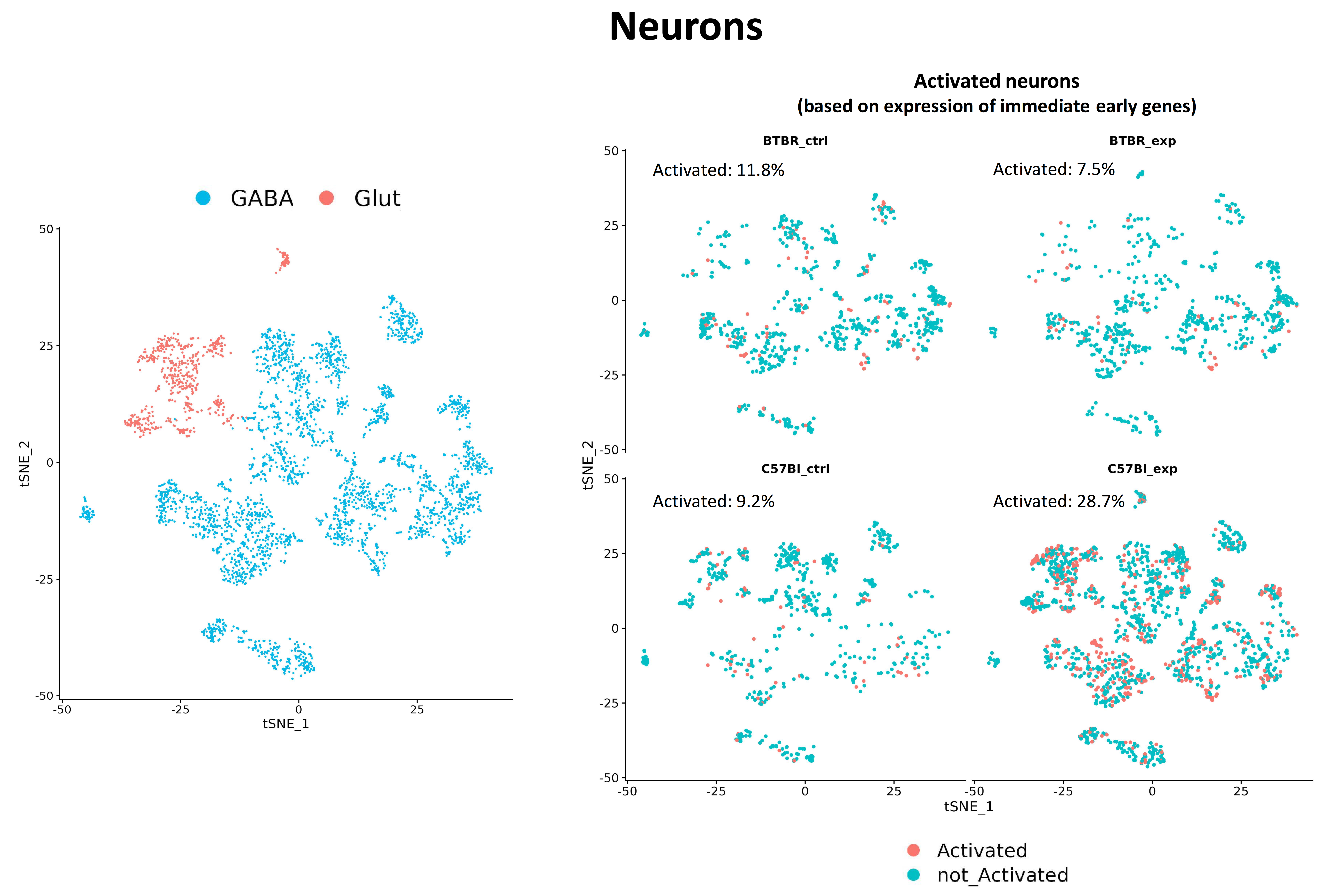
Fig. 5 Fig.5. Changes in the proportion of activated neurons in mice lines C57Bl/6 and BTBR. In wild-type mice, under experimental conditions, the proportion of activated neurons increases significantly, in contrast to BTBR mice.
Lab Members
Selected publications
Malolina, E.A., Galiakberova, A.A., Mun, V.V., Sabirov, M.S., Dashinimaev, E.B., Kulibin, A.Yu. A comparative analysis of genes differentially expressed between rete testis cells and Sertoli cells of the mouse testis. Sci Rep 13, 20896 (2023). https://doi.org/10.1038/s41598-023-48149-7
Khozyainova A.A., Valyaeva A.A., Arbatsky M.S., Isaev S.V., Iamshchikov P.S., Volchkov E.V., Sabirov M.S., Zainullina V.R., Chechekin V.I., Vorobev R.S., Menyailo M.E., Tyurin-Kuzmin P.A., Denisov E.V.. Complex Analysis of Single-Cell RNA Sequencing Data // Biochemistry (Moscow). 2023. Vol. 88, № 2. P. 231–252
Tsitrina AA, Halimani N, Andreichenko IN, Sabirov M, Nesterchuk M, Dashenkova NO, Romanov R, Bulgakova EV, Mikaelyan A, Kotelevtsev Y. 4-Methylumbelliferone Targets Revealed by Public Data Analysis and Liver Transcriptome Sequencing. Int J Mol Sci. 2023 Jan 21;24(3):2129. doi: 10.3390/ijms24032129. PMID: 36768453; PMCID: PMC9917189.
Gainullina A, Mogilenko DA, Huang LH, Todorov H, Narang V, Kim KW, Yng LS, Kent A, Jia B, Seddu K, Krchma K, Wu J, Crozat K, Tomasello E, Dress R, See P, Scott C, Gibbings S, Bajpai G, Desai JV, Maier B, This S, Wang P, Aguilar SV, Poupel L, Dussaud S, Zhou TA, Angeli V, Blander JM, Choi K, Dalod M, Dzhagalov I, Gautier EL, Jakubzick C, Lavine K, Lionakis MS, Paidassi H, Sieweke MH, Ginhoux F, Guilliams M, Benoist C, Merad M, Randolph GJ, Sergushichev A, Artyomov MN; ImmGen Consortium. Network analysis of large-scale ImmGen and Tabula Muris datasets highlights metabolic diversity of tissue mononuclear phagocytes. Cell Rep. 2023 Jan 27;42(2):112046. doi: 10.1016/j.celrep.2023.112046. Epub ahead of print. PMID: 36708514.
Klepikova A, Nenasheva T, Sheveleva O, Protasova E, Antonov D, Gainullina A, Chikina E, Sakovnich O, Gerasimova T, Nikitina I, Shevalie D, Lyadova I. iPSC-Derived Macrophages: The Differentiation Protocol Affects Cell Immune Characteristics and Differentiation Trajectories. Int J Mol Sci. 2022 Dec 16;23(24):16087. doi: 10.3390/ijms232416087. PMID: 36555728; PMCID: PMC9781144.
Dufour CR, Xia H, B'chir W, Perry MC, Kuzmanov U, Gainullina A, Dejgaard K, Scholtes C, Ouellet C, Zuo D, Sanguin-Gendreau V, Guluzian C, Smith HW, Muller WJ, Audet-Walsh E, Sergushichev AA, Emili A, Giguère V. Integrated multi-omics analysis of adverse cardiac remodeling and metabolic inflexibility upon ErbB2 and ERRα deficiency. Commun Biol. 2022 Sep 12;5(1):955. doi: 10.1038/s42003-022-03942-4. PMID: 36097051; PMCID: PMC9467976.
Emelianova M, Gainullina A, Poperechnyi N, Loboda A, Artyomov M, Sergushichev A. Shiny GATOM: omics-based identification of regulated metabolic modules in atom transition networks. Nucleic Acids Res. 2022 May 27;50(W1):W690–6. doi: 10.1093/nar/gkac427. Epub ahead of print. PMID: 35639928; PMCID: PMC9252739.